

Kasturika Shankar
Tagline:Postdoctoral Associate, University at Buffalo. Interested in understanding the fundamentals of host-pathogen interactions.
Buffalo, NY, USA
About Me
I am a scientist with a passion for understanding the molecular mechanisms underlying disease processes, particularly viral pathogenesis and its implications for human health. I earned my Ph.D. in Medical Science with an orientation towards Medical Biochemistry in May 2024 from Umeå University, Sweden, and I am currently appointed as a postdoctoral associate at the University at Buffalo, where I am studying human papillomavirus (HPV) and its role in cancer development.
My academic journey began with a Bachelor's degree in Microbiology, where I built a strong foundation in studying microorganisms and their interactions with the human body. This was followed by a Master's degree in Biotechnology, during which I honed my skills in molecular biology and developed a keen interest in the intricate mechanisms by which pathogens evade host defenses. My doctoral research further deepened my expertise as I investigated the molecular interactions between viruses and host cells, focusing on how viruses manipulate cellular machinery to evade immune responses.
During my Ph.D., I contributed significantly to the field of virology and biochemistry through ambitious research projects on enterovirus replication and protein-membrane interactions. My work resulted in several impactful publications. These achievements reflect my ability to navigate complex scientific challenges, establish protocols from scratch, and collaborate effectively within multidisciplinary teams.
Currently, my research focuses on HPV-driven cancers, such as cervical and head and neck cancers, which pose significant global health challenges, particularly in low-resource settings. Despite the availability of HPV vaccines, the incidence of HPV-related cancers continues to rise, highlighting the need for deeper insights into the viral mechanisms driving cancer progression. My work aims to unravel the complex interplay between HPV and host cellular processes, specifically how HPV manipulates translational regulation to establish persistent infections. By altering host cellular machinery, HPV ensures its survival, replication, and progression toward oncogenesis. Understanding these molecular interactions is critical for identifying therapeutic targets that can disrupt the virus’s lifecycle or halt cancer progression. Ultimately, this research seeks to contribute to novel interventions that complement existing preventive strategies and reduce the global burden of HPV-driven cancers.
In addition to my scientific expertise, I am deeply committed to mentoring and collaboration. During my Ph.D., I successfully guided undergraduate students who contributed to my publications as co-authors.
With a strong work ethic, broad training across biochemistry, biophysics, molecular biology, and cell biology, and an optimistic attitude toward tackling scientific challenges, I aim to make meaningful contributions to global health through impactful research in virology and oncology.
Education
Doctor of Philosophy - PhD
from: 2018, until: 2024Field of study:Medical biochemistry and biophysicsSchool:Umeå University
Master’s Degree
from: 2015, until: 2017Field of study:BiotechnologySchool:Indian Institute of Technology, Roorkee
Bachelor of Science - BS
from: 2012, until: 2015Field of study:MicrobiologySchool:Delhi University
Publications
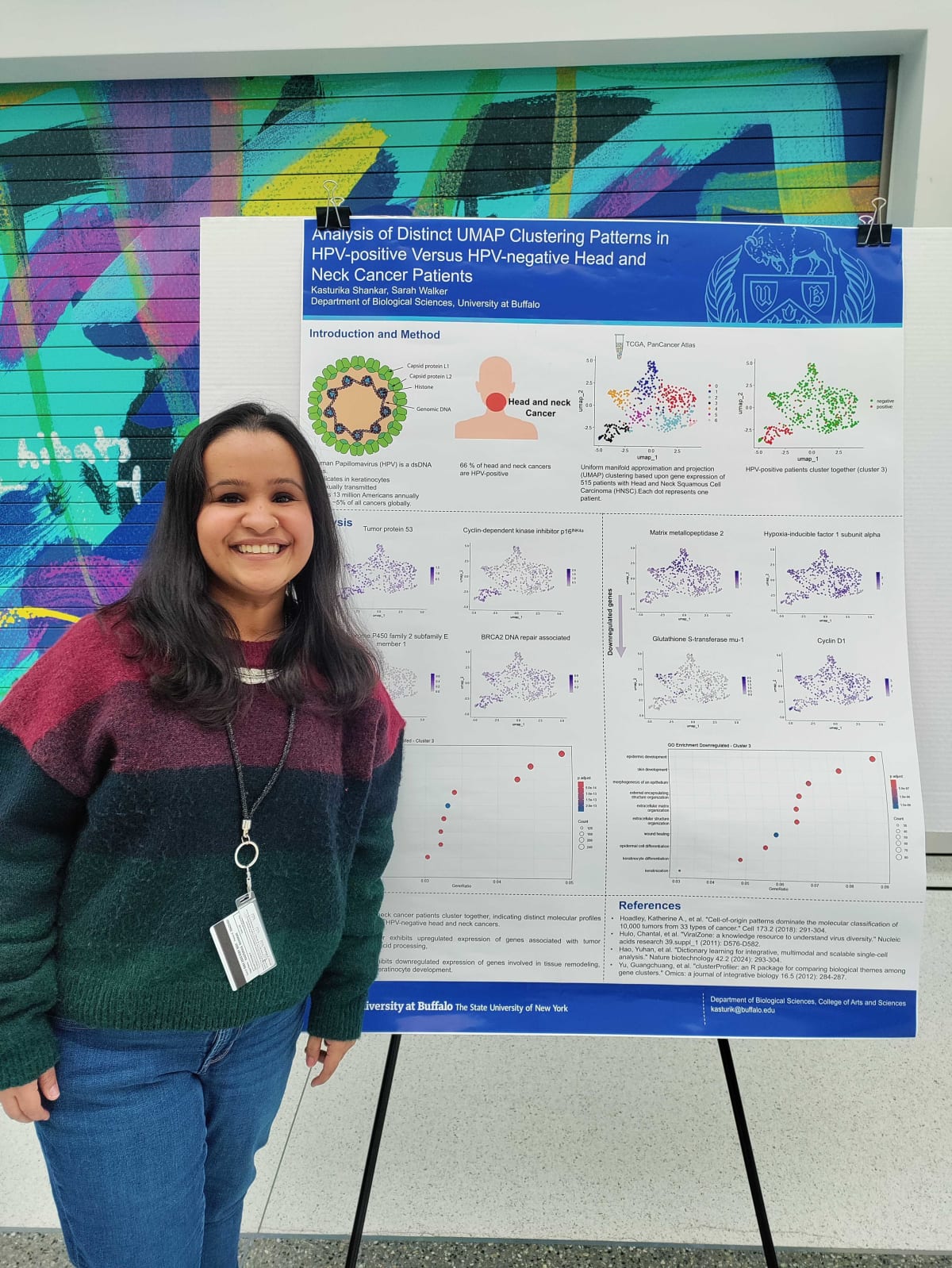
Analysis of Divergent Gene Expression between HPV+ and HPV- Head and Neck Squamous Cell Carcinoma Patients
Journal ArticleDate:2025Authors:Kasturika ShankarSarah E. WalkerDescription:Human Papillomavirus (HPV) is a non-enveloped virus with a circular double-stranded DNA genome. It is one of the most common sexually transmitted infections, with high-risk types such as HPV-16 and HPV-18 linked to anogenital and head and neck squamous cell carcinomas (HNSCC). HNSCC includes cancers of the oral cavity, pharynx, larynx, and related regions, caused by carcinogens or persistent viral infections. HPV-positive (HPV+) HNSCC cases are more prevalent in Western countries and exhibit better prognosis and treatment response compared to HPV-negative (HPV-) cases. These differences suggest distinct fundamental differences between each subtype.
This study analyzed RNA-seq data from the PanCancer Atlas 2018 dataset to investigate molecular distinctions between HPV+ and HPV-HNSCC. Using dimensionality reduction techniques such as Principal Component Analysis (PCA) and Uniform Manifold Approximation and Projection (UMAP), a clear clustering of HPV+ cases was observed, suggesting a unique gene expression profile. HPV+ tumors exhibited upregulation of genes involved in nucleic acid processing and downregulation of genes associated with apoptosis and epidermis development. These findings underscore the biological differences between HPV+ and HPV-HNSCC, offering insights into HPV-driven oncogenesis. Understanding these distinctions may improve patient stratification and inform targeted therapeutic strategies for HNSCC.
Flotation Assay With Fluorescence Readout to Study Membrane Association of the Enteroviral Peripheral Membrane Protein 2C
Journal ArticlePublisher:BIO-PROTOCOLDate:2025Authors:Kasturika ShankarYuyang LinLars-Anders CarlsonDescription:Enteroviruses are abundant pathogens of humans and animals. Their replication is strictly dependent on the conserved, viral AAA+ ATPase 2C. 2C is an oligomerizing, peripheral membrane protein, and its low solubility as recombinant protein has hampered functional studies of the full-length, recombinant protein bound to a membrane. Here, we describe a modification of the classical, ultracentrifugation-based liposome flotation assay optimized to study the interaction of recombinant 2C with membranes and the functions of membrane-bound, full-length recombinant 2C. The assay takes advantage of the high solubility of recombinant 2C while fused to a maltose-binding protein. Removing this solubility-enhancing tag by specific protease cleavage in the presence of liposomes allows 2C to associate with membranes prior to aggregating. Fluorophore labeling of protein and liposomes allows rapid and precise quantitation of 2C’s association with membranes. This assay is adaptable to any peripheral membrane protein that can be fluorophore-labeled and expressed as a solubility-enhancing fusion protein.
Biochemical and structural studies of proteins supporting the genome replication of enteroviruses and Giardia intestinalis
ThesisPublisher:Umeå UniversityDate:2024Authors:Description:The Enterovirus genus of the Picornaviridae family includes non-enveloped, positive-sense single-stranded RNA (ssRNA) viruses. This genus of viruses causes many diseases such as poliomyelitis by poliovirus (PV), cardiomyopathy by coxsackievirus B3 (CVB3), common cold by rhinoviruses (RVs) and meningitis by Enterovirus 71 (EV 71). The 7.5 kb enterovirus genome encodes a polyprotein, which is subsequently cleaved to yield viral proteins. These viral proteins hijack and modify the membranes of the Golgi and ER to give rise to replication organelles (ROs).
In the first paper of my thesis, we showed that membranes of the ROs act as an assembly line for the assembly of enteroviruses. Using cryo-electron tomography we studied poliovirusinfected cells. The one feature that we found most interesting was that a protein was tethering viral capsid to membranes as well as two membranes together. We hypothesized that viral protein 2C was a strong candidate. Therefore, I tried to mimic the interaction between 2C and viral capsid using purified Poliovirus and 2C protein. However, after trying several biochemical conditions I could not mimic this interaction. This strongly indicated that I might need to have a membrane in the system and this led me to study the membrane binding activity of 2C in the second paper.
In the second paper, I studied an Enteroviral multi-functional protein: 2C, a highly conserved AAA+ ATPase that plays an important role in the biogenesis of ROs and virus assembly. One of the most interesting features of 2C is its N-terminal membrane-binding domain consisting of 40 amino acids. Using in vitro reconstitution methods and biochemistry, I investigated the association of full-length 2C with lipid vesicles and how this affects the function of the protein. I showed that amino acids 12 to 40 are important not only for membrane binding but also for hexamerization. Moreover, truncation of the first 11 amino acids leads to loss of membrane tethering activity of the protein, which is essential for the formation of ROs. I was also able to demonstrate that the protein is sufficient to recruit RNA to the membrane as it was not previously known how RNA replication is localized to the membrane and here, we found the possible mechanism. In this realistic reconstituted system, I have shown that 2C is not a helicase but an ATP-independent RNA chaperone. Collectively, these discoveries offer a biochemical foundation for various functions of 2C in enterovirus replication. This sets up a more practical biochemical framework for future research, which could potentially contribute to the development of drugs aimed at thwarting enterovirus infections.
In the third project, I studied deoxyadenosine kinase (dAK) from Giardia intestinalis, a protozoan responsible for severe diarrhea spread by the fecal-oral route. Giardia is completely dependent on the host for deoxyribonucleosides as it lacks a de novo pathway for their synthesis. dAK uses ATP as a phosphate donor to phosphorylate deoxyadenosine, this activity is essential for genome replication of the protozoan. In this project, we have biochemically and structurally characterized dAK. Here, I used cryo-electron microscopy (cryo-EM) single particle analysis to show that dAK exists as a homo-tetramer in solution. This is an important finding because this is the first example of a non-thymidine kinase1-like deoxyribonucleoside kinase with a tetrameric structure and subsequent mutagenesis analysis showed that tetramerization allows the enzyme to achieve a higher affinity for its deoxyadenosine substrate.
In summary, in my thesis, I have biochemically and structurally characterized proteins supporting the genome replication of enteroviruses and Giardia intestinalis.
A chemical inhibitor of IST1-CHMP1B interaction impairs endosomal recycling and induces noncanonical LC3 lipidation
Journal ArticlePublisher:Proceedings of the National Academy of SciencesDate:2024Authors:KnyazevaAnastasiaShuang LiDale P. CorkeryKasturika ShankarLaura K. HerzogXuepei ZhangBirendra Singh et al.Description:A group of proteins, the endosomal sorting complex required for transport (ESCRT), plays a crucial role in membrane remodeling—a cellular process involving the rearrangement of cellular membranes. Understanding and controlling specific parts of ESCRT is challenging due to its complex organization. Here, we describe a natural product-like compound Tantalosin that specifically disrupts a particular part of ESCRT called the IST1-CHMP1B complex. Using Tantalosin as a chemical tool, we examine how this complex contributes to different membrane remodeling events. Interestingly, Tantalosin induces noncanonical LC3 lipidation—a process involving noncanonical autophagy (self-eating)—at stalled endosomes. This finding further enriches our understanding of ESCRT function and highlights the importance of small molecules as valuable tools for unraveling complex biological mechanisms.
Tetramerization of deoxyadenosine kinase meets the demands of a DNA replication substrate challenge in Giardia intestinalis
Journal ArticlePublisher:Nucleic Acids ResearchDate:2024Authors:Farahnaz RanjbarianKarim RafieKasturika ShankarSascha KrakovkaStaffan G SvardLars-Anders CarlsonAnders HoferDescription:The protozoan parasite Giardia intestinalis is one of only a few organisms lacking de novo synthesis of DNA building blocks (deoxyribonucleotides). Instead, the parasite relies exclusively on salvaging deoxyadenosine and other deoxyribonucleosides from its host environment. Here, we report that G. intestinalis has a deoxyribonucleoside kinase with a 1000-fold higher catalytic efficiency (kcat/KM) for deoxyadenosine than the corresponding mammalian kinases and can thereby provide sufficient deoxyadenosine triphosphate levels for DNA synthesis despite the lack of de novo synthesis. Several deoxyadenosine analogs were also potent substrates and showed comparable EC50 values on cultured G. intestinalis cells as metronidazole, the current first-line treatment, with the additional advantage of being effective against metronidazole-resistant parasites. Structural analysis using cryo-EM and X-ray crystallography showed that the enzyme is unique within its family of deoxyribonucleoside kinases by forming a tetramer stabilized by extended N- and C-termini in a novel dimer–dimer interaction. Removal of the two termini resulted in lost ability to form tetramers and a markedly reduced affinity for the deoxyribonucleoside substrate. The development of highly efficient deoxyribonucleoside kinases via oligomerization may represent a critical evolutionary adaptation in organisms that rely solely on deoxyribonucleoside salvage.
In vitro reconstitution reveals membrane clustering and double-stranded RNA recruitment by the enteroviral AAA+ ATPase 2C
Journal ArticlePublisher:Plos PathogensDate:2024Authors:Kasturika ShankarMarie N. SorinHimanshu SharmaOskar SkoglundSelma DahmaneJosy ter BeekSolomon TesfalidetLouise NenzenLars-Anders CarlsonDescription:Enteroviruses are frequent causes of diseases that range from mild common colds to severe conditions such as poliomyelitis, acute flaccid myelitis and viral myocarditis. Enterovirus particles are tiny, even by virus standards, and they can only package a small amount of viral genetic material. With such a small genetic ‘toolbox’, how can these viruses so vigorously hijack the interior of a cell and turn it into a viral factory? One reason is that the viral genes often encode multifunctional proteins. Here, we studied one such multifunctional viral protein, called 2C. This protein is found in all enteroviruses and in related viruses, and the virus cannot replicate without it. We developed a new way of studying 2C that allowed us to work with the complete 2C protein ‘in its element’, namely bound to a membrane. We found that 2C not only binds one membrane, but can gather several membranes together–a function that may be important to building the viral factory. We also found that 2C can bind RNA (the chemical form of the virus’ genetic material) to membranes and can cleave RNA. Our findings reinforce the view of 2C as an organizer of the viral factory.
Membrane-assisted assembly and selective secretory autophagy of enteroviruses
Journal ArticlePublisher:Nature CommunicationsDate:2022Authors:Selma DahmaneAdeline KervielDustin R. MoradoKasturika ShankarBjörn AhlmanMichael LazarouNihal Altan-BonnetLars-Anders CarlsonDescription:Enteroviruses are non-enveloped positive-sense RNA viruses that cause diverse diseases in humans. Their rapid multiplication depends on remodeling of cytoplasmic membranes for viral genome replication. It is unknown how virions assemble around these newly synthesized genomes and how they are then loaded into autophagic membranes for release through secretory autophagy. Here, we use cryo-electron tomography of infected cells to show that poliovirus assembles directly on replication membranes. Pharmacological untethering of capsids from membranes abrogates RNA encapsidation. Our data directly visualize a membrane-bound half-capsid as a prominent virion assembly intermediate. Assembly progression past this intermediate depends on the class III phosphatidylinositol 3-kinase VPS34, a key host-cell autophagy factor. On the other hand, the canonical autophagy initiator ULK1 is shown to restrict virion production since its inhibition leads to increased accumulation of virions in vast intracellular arrays, followed by an increased vesicular release at later time points. Finally, we identify multiple layers of selectivity in virus-induced autophagy, with a strong selection for RNA-loaded virions over empty capsids and the segregation of virions from other types of autophagosome contents. These findings provide an integrated structural framework for multiple stages of the poliovirus life cycle.
A 3D view of how enteroviruses hijack autophagy
ReviewPublisher:AutophagyDate:2022Authors:Selma DahmaneKasturika ShankarLars-Anders CarlsonDescription:Viruses are masters at using cellular pathways to aid their replication. Cryo-electron tomography of poliovirus-infected cells revealed how it utilizes macroautophagy to its advantage. Assembly of these non-enveloped virions takes place directly on membranes and requires PIK3C3/VPS34 activity to be completed, whereas the canonical autophagy inducer ULK1 restricts virus assembly. The tomograms further revealed that enterovirus-induced autophagy is selective for RNA-loaded virions, which may help ensure maximum infectivity of the virus-laden vesicles released through secretory autophagy.
Work Experiences
Postdoctoral Associate
from: 2024, until: presentOrganization:University at Buffalo
PHD Student
from: 2018, until: 2024Organization:Umeå UniversityLocation:Umea, Sweden
Junior Research Fellow
from: 2018, until: 2018Organization:Indian Institute of Technology, Roorkee
Skills
- Structural Biology
- Microbiology
- Molecular & Cellular Biology
- Biochemistry
- Cryogenic Electron Microscopy
- Molecular Virology
- University teaching and mentoring